Automated Endoleak and AAA Detection
This project represents more of a series of incremental projects that I worked on starting in the Summer of 2018 up until Spring 2019. This project represented a new collaboration between the Complex Systems Center, which I has just joined as a Master’s student in Safwan Wshah’s lab, and the UVM medical center. On the UVM medical center end of things, I worked mainly with two excellent doctors, vascular surgeon Daniel Bertges and interventional radiologist Christopher Morris.
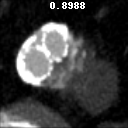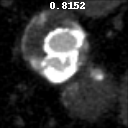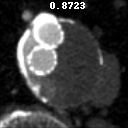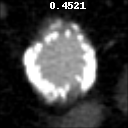
The goal of this project was to build from the ground up a comprehensive set of tools to assist, in an automated way, in various tasks related to the pathology around Abdominal Aortic Aneurysm (AAA) patient care. In particular, we developed tools to first isolate a smaller region of interest around the AAA, perform EndoLeak detection and lastly 3D segmentation of the Aneurysm itself (providing automated measurements of AAA volume). The input to all of these tools was computed tomography angiography (CTA) imaging from a collection of patients who underwent care at the University of Vermont Medical center.
This project was made both more interesting and difficult in that no pre-existing labelled dataset existed. In this way it differs greatly from simmilar projects on prepared public datasets for say the prediction of lung cancer. In this project we instead had to work from the level of coarse natural text labelling that existing as saved in the UVM medical center. We were therefore required to build the deep learning system through a series of creative semi-automated labelling schemes, as well as semi-automated text mining tools. In particular, sparse 3D labelling was used for the by far most intenstive piece (3D dense segmentation of AAA).
A paper on this work “Machine deep learning accurately detects endoleak after endovascular abdominal aortic aneurysm repair” was published in JVS-Vascular Science. This paper recieved the 2020 JVS most viewed article award. This work was also presented as a keynote presentation at the 2019 Vascular Annual Meeting of the Society for Vascular Surgery, National Harbor, Md, June 12-15, 2019. An earlier, more methods focused, paper Deep Learning for Recognition of Endoleak After Endovascular Abdominal Aortic Aneurysm Repair was published and presented as a poster at the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). Lastly, this worked was filled in US Patent Application No.: 17/602,164 Title: METHOD AND APPARATUS FOR ANALYZING AORTIC ANEURYSMS AND ENDOLEAKS IN COMPUTED TOMOGRAPHY SCANS.
Due to various difficulties related to patient privacy, only a generalized subset of the code is avaliable for this project on github.